脊髓损伤
-
Figure 1|Establishment of zebrafish SCI model and evaluation of motor function after SCI.
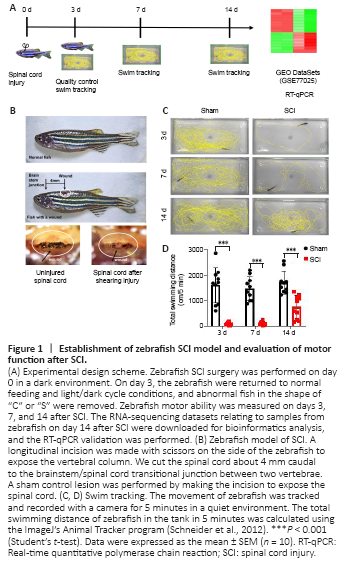
The subacute phase is critical to zebrafish spinal cord recovery, when glial bridge formation and spinal cord regeneration occur (Mokalled et al., 2016). Therefore, the 2-week time point was used to investigate the mechanisms controlling locomotor recovery. The study design is presented in Figure 1A. First, a zebrafish SCI model was produced, and sham surgery was used as the control (Figure 1B). Zebrafish with “C” and “S” shapes were considered to have damaged spines and were excluded on day 3 after SCI to ensure the consistency of SCI surgery results. Motor function of zebrafish was observed on days 3, 7, and 14 after SCI (Figure 1C and D). The results show that zebrafish did not recover at 3 days or 7 days after SCI. They recovered 44% of their swimming ability 14 days after SCI (P < 0.001).
Figure 2|Differentially expressed genes 2 weeks after SCI in zebrafish.

To target the genes that play the most important role after SCI, we first obtained the DEGs after SCI by using DESeq2 (|log2FoldChange| > 0.5 and FDR < 0.001). In total, 7762 DEGs were identified after SCI, including 2950 up-regulated and 4812 down-regulated DEGs. We sorted the DEGs from highest-to-lowest-expressed genes in the injury group and obtained the top 410 higher expressed genes (DHEGs). Finally, we obtained the top 50 up-regulated and down-regulated DHEGs (Figure 2), which are also strongly expressed. We predicted these genes would play important roles in SCI repair.
Figure 3|GO and KEGG pathway enrichment of differentially expressed genes.
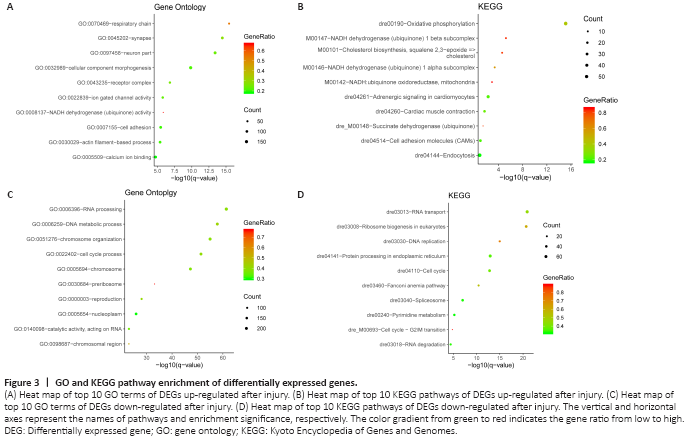
DEGs were characterized by GO and KEGG enrichment using Metascape. The top 10 pathways that reached a statistically significant value (P < 0.05) are shown in Figure 3 and Tables 2 and 3. For the GO analysis, the up-regulated genes are mainly enriched in respiratory chain, synapse, neuron part, cellular component morphogenesis, and Ca2+-binding pathways. The up-regulated DEGs relate to neuronal repair and regeneration. The down-regulated genes are mainly enriched in RNA processing, DNA metabolic process, chromosome organization, and cell cycle. For the KEGG pathway analysis, the signaling pathways of up-regulated genes are mainly enriched in oxidative phosphorylation, NADH dehydrogenase (ubiquinone) pathway, and endocytosis. The signaling pathways of down-regulated differential genes are mainly enriched in RNA transport, protein processing in endoplasmic reticulum, and cell cycle. The GO and KEGG analyses of down-regulated DEGs show that these genes are mainly involved in cellular replication.
Figure 4|Distribution of the genes into the GO pathways.
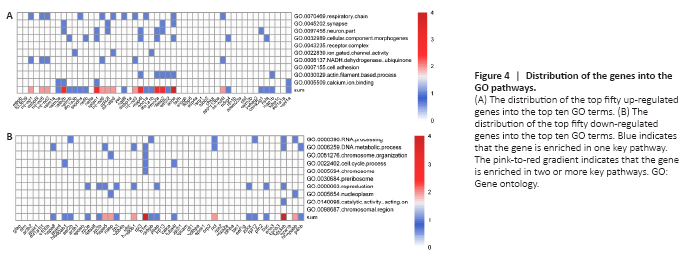
Through GO analysis, we found that the up-regulated genes are enriched in synapses and neurons. Hence, we assumed that up-regulated genes are beneficial for SCI repair. We distributed the top 50 genes into the top 10 GO pathways to determine how the top 50 up-regulated genes perform certain functions and to identify the most important genes after SCI (Figure 4A). Most of the top 30 of the top 50 genes fall into at least one pathway. Furthermore, calb2b, sptan1, map4l, stmn2a, clasp2, and ache fall into at least three pathways, which implies that these genes may play key roles in their respective pathways. These genes are related to the components and repair of neurons, such as respiratory chain, synapse, neuron part, and Ca2+ binding. We also distributed the top 50 down-regulated genes into the top 10 GO pathways. The results showed that h1m and ddx3xb fall into four pathways (Figure 4B), and all the genes are involved in cellular replication.
Figure 5|RT-qPCR verified partial DEGs and PPI.
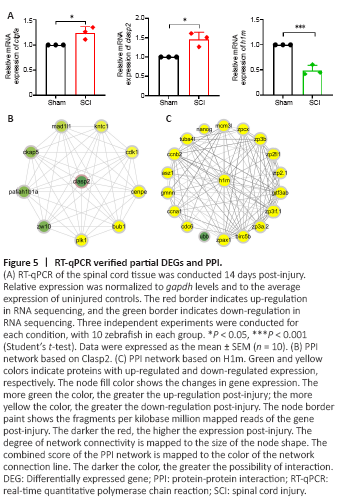
Key genes in Figure 4A and Figure 4B are involved in multiple pathways, suggesting that they play an important role in spinal cord repair. Therefore, we performed RT-qPCR to verify the expression changes of calb2b, sptan1, map41, stmn2a, clasp2, ache, h1m, and ddx3xb after SCI. The mRNAs encoding calb2b, sptan1, map41, stmn2a, clasp2, and ache are up-regulated, and the mRNAs encoding h1m and ddx3xb are down-regulated in the RNA sequencing dataset for SCI. According to Mokalled et al. (2016), ctgfa is up-regulated 2 weeks after zebrafish spinal cord crush injury (P < 0.05). Ctgfa was used as a positive control for this dataset, and it was also up-regulated consistently, as previously reported (Mokalled et al., 2016) (Figure 5A). RT-qPCR results verified that clasp2 is up-regulated (1.46 times, P < 0.05) and h1m is down-regulated (0.46 times, P < 0.001) after SCI (Figure 5A). The expression changes are consistent with the RNA sequencing (RNAseq) transcriptome data. The RT-qPCR results show that sptan1 and stmn2a were unchanged and calb2b, map4l, and ache were downregulated to 0.48, 0.33, and 0.62 times the sham group, respectively, and ddx3xb was up-regulated to 1.69 times the sham group (Additional Figure 2).
To further predict the specific molecular mechanisms of Clasp2 and H1m for regulating nerve injury repair-related pathways after SCI, protein-protein interaction networks were created for Clasp2 protein or H1m protein using the STRING database version 11.0 (https://string-db.org/). PPI network analysis showed the interactions among the DEGs. There were nine DEGs interacting with Clasp2 (Figure 5B). Pafah1b1a (Drerup et al., 2010) is involved in the development of the nervous system, and Ckap5 is associated with microtubule dynamics (Cassimeris et al., 2009; Chanez et al., 2015). There were 18 DEGs interacting with H1m (Figure 5C); most of their functions are related to cell cycle and embryonic development, such as Cdc6 (Yao et al., 2017), Ccna1 (Beaudoin et al., 2018), and Nanog (He et al., 2020; Palfy et al., 2020).
Figure 6|Schematic map of the involvement of key genes in the two facets of spinal cord regeneration.
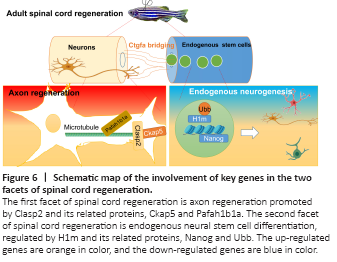
Microtubules play an important role in the formation and maintenance of axons and dendrites in neurons. Clasp2 is a microtubule plus-end tracking protein that promotes the stabilization of dynamic microtubules (Maki et al., 2015). Because it is induced in zebrafish upon SCI, we hypothesized that it has a pro-regenerative role. Clasp2 mediates growth-cone microtubules and is in exquisite balance with the co-regulation of GSK3 (Hur et al., 2011). It has also been reported that Clasp2 regulates neuronal polarity and synaptic function, and it has been found that overexpression of Clasp2 can lead to an increase in presynaptic-terminal circumference and total synapse number (Beffert et al., 2012). Dillon et al. (2017) demonstrated that Clasp2 regulates neuronal production and controls neuronal migration in mice. Through PPI network analysis, we found that Clasp2 interacts with microtubule-related proteins. Ckap5 plays an important role in axon growth and regulates the growth cone by promoting interaction between microtubules and F-actin (Lowery et al., 2013; Slater et al., 2019). Pafah1b1a (also known as lissencephaly-1 or LIS1) activates dynein, which facilitates microtubule transportation that is critical for axon regeneration (Elshenawy et al., 2020; Htet et al., 2020; Marzo et al., 2020; McKenney, 2020). We hypothesized that Clasp2 may promote the growth of neuronal axons and growth cones (Figure 6). Neurogenesis is a key process in zebrafish spinal cord regeneration (Ceci et al., 2018). Neurogenesis may come from transdifferentiation as well as from direct differentiation from neural stem cells or progenitor cells. Notwithstanding transcription factors, non-coding RNAs, such as microRNAs, play an emerging role in transdifferentiation (Sabater et al., 2020). Therefore, further mining of the RNAseq data for the microRNAome or investigating single-cell RNAseq techniques are needed to study neurogenesis.