颅神经损伤
-
Figure 1|SB216763 suppresses DNA damage and increases cell viability in serum-deprived retinal neurons.
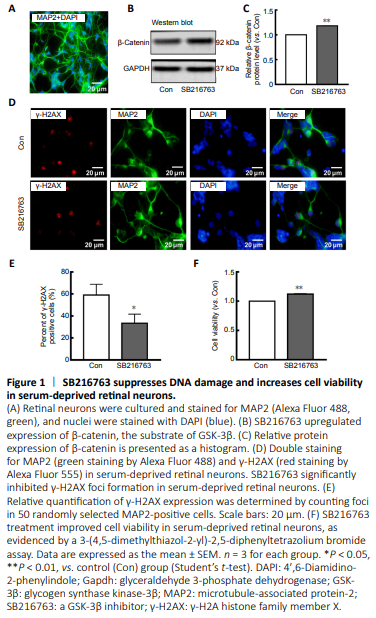
Many studies have indicated that GSK-3β inhibition protects neurons from apoptosis induced by genotoxic and other stresses (Marchena et al., 2017). To explore the underlying mechanism, primary rat retinal neurons were cultured and subjected to serum starvation. Figure 1A shows that 92% of primary cultured retinal cells were positive for MAP2, a well-known neuronal marker (Johnson and Jope, 1992), demonstrating their neuronal characteristics. Pretreatment of primary rat retinal neurons for 24 hours with SB216763, a GSK-3β inhibitor, prior to serum starvation resulted in upregulation of β-catenin, a downstream target negatively regulated by GSK-3β (P < 0.01; Figure 1B and C). These results confirmed that SB216763 mediated GSK-3β inhibition in ischemic retinal neurons. Relative intensities of bands were quantified by densitometry and normalized to GAPDH levels. As shown in Figure 1D, double staining of MAP2 (a neuronal marker) and γ-H2AX [a well-characterized DNA DSB marker (Ivashkevich et al., 2012)] demonstrated that serum deprivation induced pronounced γ-H2AX foci formation in retinal neurons; however, inhibiting GSK-3β with SB216763 pretreatment significantly decreased γ-H2AX expression compared with the control group (P < 0.05; Figure 1E). Accordingly, SB216763 pretreatment significantly improved the cell viability of retinal neurons upon serum deprivation compared with the control treatment (DMSO) (P < 0.001; Figure 1F). These results indicate that GSK-3β inhibition suppressed DNA damage and promoted cell viability in retinal neurons suffering from ischemic injury. However, the mechanism by which GSK-3β inhibition mediates DNA repair is still unknown.
Figure 2|Ligase IV is involved in the neuroprotective mechanisms of SB216763.

LiCl has been used as a GSK-3 inhibitor in clinical treatment and scientific research for many years. Our previous study indicated that LiCl increased DNA stability by upregulating DNA ligase IV, thereby facilitating neuroprotective effects (Zhuang et al., 2009; Yang et al., 2011). Therefore, we hypothesized that the GSK-3β inhibitor SB216763 might regulate DNA ligase IV. Accordingly, we measured mRNA and protein levels of DNA ligase IV in primary retinal neurons treated with SB216763 or vehicle control. Consistent with our expectations, 24-hour treatment with SB216763 (5 μM) significantly increased mRNA expression of ligase IV in retinal neurons suffering from ischemic injury compared with control cells pretreated with DMSO vehicle (P < 0.001; Figure 2A). The same effect was observed for protein expression of ligase IV in ischemic retinal neurons (P < 0.01; Figure 2B and C).
To further confirm whether ligase IV was involved in SB216763-mediated neuroprotection in retinal neurons suffering from ischemic injury, ligase IV expression was silenced using siRNA interference. As shown in Figure 2D, siRNA interference suppressed the upregulation of ligase IV induced by SB216763 in retinal neurons under serum deprivation conditions (P < 0.05; Figure 2E). Accordingly, SB216763 significantly improved the cell viability of retinal neurons upon serum deprivation, while this increase was abrogated by ligase IV siRNA interference (P < 0.05; Figure 2F). These results support the potential involvement of ligase IV expression in the neuroprotective effect of SB216763.
Figure 3|SB216763 is involved in transcriptional regulation of ligase IV in retinal neurons.
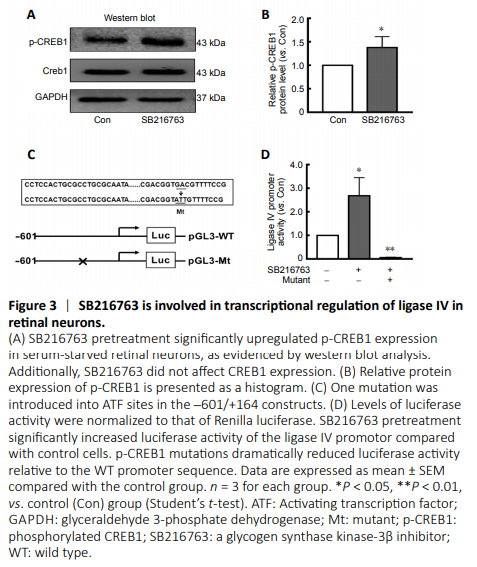
Our previous study reported that p-CREB1 may participate in the transcriptional regulation of ligase IV (Yang et al., 2016). Here, we found that SB216763 did not affect CREB1 expression, but instead markedly upregulated p-CREB1 expression in retinal neurons following serum deprivation (Figure 3A). Indeed, SB216763 increased CREB1 phosphorylation by 1.38-fold over the control treatment (P < 0.05; Figure 3B).
The transcriptional mechanism of ligase IV was further investigated by a luciferase reporter assay using a construct driven by the ligase IV proximal promoter (–601 to +164), which contains an activating transcription factor binding site between –121 and –301 (Yang et al., 2016). The transcription factor binding motif of the p-CREB1 DNA-binding sequence (5′-GTG ACG TT-3′) is located at –126. As shown in Figure 3C, the p-CREB1 site was mutated (5′-GTG CCG TT-3′ to 5′-GTA TTG TT-3′) and the entire promoter region from –601 to +164 was assayed in ischemic retinal neurons pretreated with 5 μM SB216763. Twenty-four hours after pretreatment with SB216763, serum-starved retinal neurons were transfected with luciferase reporter constructs containing the ligase IV promoter. At 24 hours post-transfection, levels of firefly and Renilla luciferase activity were measured using the Dual-Glo Luciferase Assay System. The results indicated that SB216763 pretreatment significantly increased luciferase activity compared with control cells. However, p-CREB1 mutations dramatically reduced luciferase activity relative to that of the wild-type promoter sequence (P < 0.05; Figure 3D). These results further suggest that inhibiting GSK-3β elicits neuroprotection by regulating ligase IV expression in ischemic retinal neurons.
Figure 4|GSK-3β activity is inhibited by different mechanisms in retinal neurons.

GSK-3β activity is regulated by phosphorylation of Tyr216 or Ser9 (Park et al., 2013). However, SB216763 and LiCl, both well-known GSK-3β inhibitors, have different effects on Ser9 phosphorylation of GSK-3β in various cell lines (Zhang et al., 2003; Noble et al., 2005; Koo et al., 2014; D’Angelo et al., 2016). To explore what occurs in retinal neurons, we further investigated GSK-3β expression and phosphorylation levels in retinal neurons treated with LiCl and SB216763. The relative intensities of bands were quantified by densitometry and normalized to GAPDH levels. As shown in Figure 4A, LiCl did not affect GSK-3β expression in ischemic retinal neurons compared with the control treatment (P > 0.05; Figure 4A and B). However, LiCl pretreatment significantly upregulated p-GSK-3β expression in ischemic retinal neurons (P < 0.05; Figure 4A and B), indicating that LiCl indirectly inhibited GSK-3β activity by stimulating GSK-3β phosphorylation in retinal neurons. In contrast, SB216763 not only directly decreased GSK-3β expression (P < 0.05), but also dramatically inhibited its phosphorylation (P < 0.001; Figure 4C and D).
Figure 5|SB216763 protects retinal neurons from ischemic injury in vivo.

To further confirm the neuroprotective mechanism of SB216763, a rat I/R model was established to mimic retinal ischemic injury in vivo. As shown in Figure 5A, SB216763 pretreatment significantly upregulated the expression of both ligase IV (P < 0.05) and p-CREB1 (P < 0.01; Figure 5B) in ischemic rat retinas. These observations were consistent with those of our in vitro studies. Moreover, we analyzed γ-H2AX foci formation at the indicated time points (1 and 3 days). Our results showed that γ-H2AX-positive cells were primarily located in the retinal ganglion cell layer after I/R treatment (1 day; Figure 5C). After counting the number of γ-H2AX foci in at least 10 slides, we found that SB216763 pretreatment significantly suppressed γ-H2AX foci formation in the ganglion cell layer at 1 day (P < 0.05) and 3 days (P < 0.001) following I/R surgery (Figure 5D). Thus, these data further indicate that the GSK-3β inhibitor SB216763 enhanced repair of DNA DSBs by upregulating ligase IV expression in serum-deprived retinal neurons.