NRR:青岛大学李菁团队提出支架蛋白CNKSR2在神经发育障碍的潜在机制
撰文:尹琳
RAS激酶抑制因子连接增强蛋白2(connector enhancer of kinase suppressor of Ras 2, CNKSR2)是一种具有多个蛋白结合结构域的支架蛋白,在神经细胞的突触后致密组分(postsynaptic density, PSD)及细胞质中均有分布,其基因变异与智力障碍等神经发育性疾病相关,但致病机制尚不清楚。研究发现,CNKSR2蛋白可协助PSD蛋白复合物的组织和膜相关定位,影响突触后信号传导和树突棘形态发生;然而,对于细胞质中CNKSR2蛋白的功能还有待阐明。
最近,来自中国青岛大学李菁团队在《中国神经再生研究(英文)》(Neural Regeneration Research)上发表了题为“CNKSR2 interactome analysis indicates its association with the centrosome/microtubule system”的研究。该研究通过对CNKSR2相互作用蛋白的鉴定和分析,发现这些蛋白显著富集于中心体蛋白质组中。然后通过免疫荧光实验验证了CNKSR2与中心体蛋白CEP63的共定位。继而在小鼠脑神经瘤细胞系N2A细胞中下调CNKSR2的表达,可见一系列中心体相关基因的表达受到影响,同时细胞形态、增殖及迁移能力等中心体相关功能也发生改变。将CNKSR2互作组与神经发育性疾病相关基因进行比对发现,CNKSR2互作蛋白显著富集于智力障碍和孤独症谱系障碍相关基因中。研究揭示了CNKSR2与中心体之间的联系,提出Cnksr2变异可能通过破坏中心体结构完整性,从而引起神经细胞功能失调的科学假说,这丰富了对于CNKSR2的认识,为神经发育性疾病致病机制的探索提供了新的视角。
CNKSR2广泛分布于细胞内,定位于细胞质膜相关结构和细胞质成分中,该研究用含不同去垢剂(脱氧胆酸钠和Triton-X100)的裂解液提取蛋白质进行免疫共沉淀实验,结合质谱分析鉴定CNKSR2的互作蛋白,并利用在线生物信息学平台Metascape(https://metascape.org)和STRING (https://string-db.org/)对这些蛋白进行了生物学过程和分子功能分析,发现CNKSR2互作蛋白与微管组织密切相关(图1)。
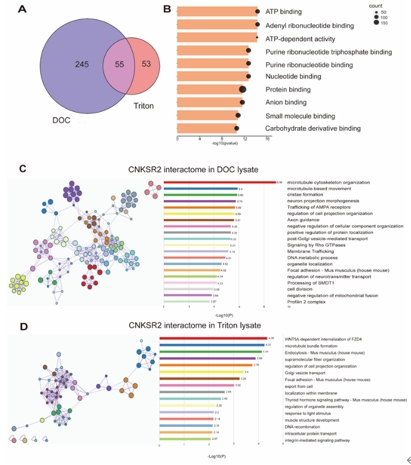
图1 CNKSR2 相互作用组概览(图源:Yin et al., Neural Regen Res, 2025)
李菁等利用从文献和数据库中收集到的重要细胞器的蛋白质组数据,对CNKSR2互作蛋白进行了富集分析,结果表明,CNKSR2互作蛋白在PSD和中心体蛋白质组中显著富集。使用免疫共沉淀-免疫印迹相结合的方法,验证了CNKSR2与微管蛋白DYNC1H1及中心体蛋白CEP290的相互作用,并通过免疫荧光的方法证实CNKSR2与中心体蛋白CEP63的共定位(图2)。
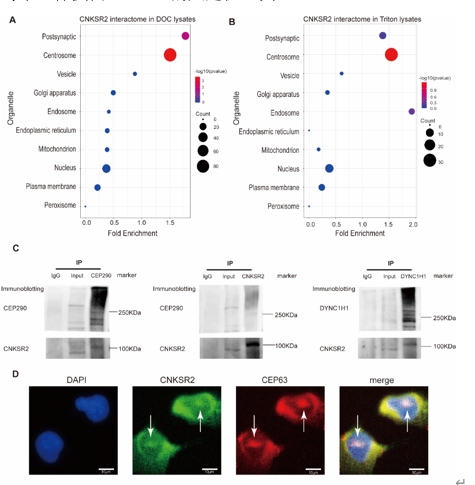
图2 CNKSR2 与中心体相互作用组的关联(图源:Yin et al., Neural Regen Res, 2025)
中心体由几个具有不同功能的特定区域组成。最近一项关于中心体空间蛋白质组的研究通过靶定中心体亚结构的标志性蛋白,分析了中心体不同区域的蛋白组成[1]。李菁团队将鉴定到的CNKSR2互作蛋白与这些数据集对比,绘制了CNKSR2互作蛋白的中心体亚结构分布图(图3),其显示CNKSR2互作组与中心体近端结构关系更为密切。

图3 CNKSR2互作组的亚中心体分布分析(图源:Yin et al., Neural Regen Res, 2025)
下调CNKSR2表达会影响中心体相关功能,包括细胞形态、细胞增殖及运动能力(图4)。下调CNKSR2后许多中心体相关基因表达降低。

图4下调CNKSR2表达影响中心体相关功能(图源:Yin et al., Neural Regen Res, 2025)
将CNKSR2互作组与已报道的神经发育疾病相关基因[2-8]进行比对分析,结果显示CNKSR2互作蛋白在孤独症谱系障碍相关基因群组中显著富集(图5)。
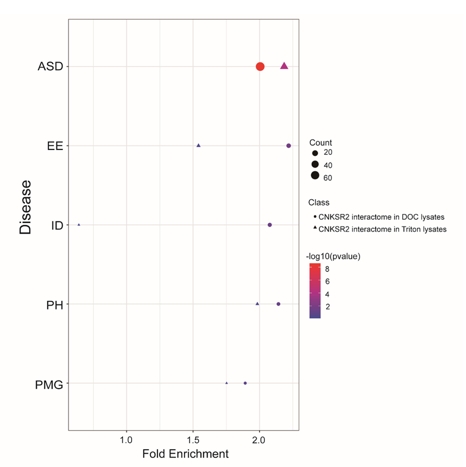
图5 CNKSR2互作组在神经发育疾病相关基因中的富集分析(图源:Yin et al., Neural Regen Res, 2025)
总的来说,此研究通过鉴定和分析CNKSR2的相互作用蛋白,发现CNKSR2与许多中心体/微管蛋白相关;下调CNKSR2表达影响许多中心体相关蛋白的表达,并影响N2A细胞的形态、增殖和运动能力;对CNKSR2互作组与神经发育疾病相关基因数据集比对发现CNKSR2互作蛋白显著富集于孤独症谱系障碍相关基因中。该研究揭示了CNKSR2与中心体之间的联系,提出CNKSR2变异可能通过破坏中心体结构完整性,影响中心体相关功能,从而引起神经细胞功能失调的科学假说,为探索与CNKSR2相关的致病机制提供了新的线索。
然而,这项研究尚无法确定CNKSR2在中心体上的直接相互作用蛋白,因此也未能进一步研究CNKSR2在中心体组织和功能中的具体作用和分子机制,在这一方面还有待未来更加深入的探索。
原文链接:https://doi.org/10.4103/NRR.NRR-D-23-01725
参考文献
[1] O'neill AC, Uzbas F, Antognolli G, et al. Spatial centrosome proteome of human neural cells uncovers disease-relevant heterogeneity. Science. 2022;376(6599):eabf9088.
[2] De Ligt J, Willemsen MH, Van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367(20):1921-1929.
[3] Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380(9854):1674-1682.
[4] Hamdan FF, Srour M, Capo-Chichi JM, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10(10):e1004772.
[5] Lim ET, Uddin M, De Rubeis S, et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci. 2017;20(9):1217-1224.
[6] Epi4k Consortium, Epilepsy Phenome/Genome Project, Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217-221.
[7] Heinzen EL, O'neill AC, Zhu X, et al. De novo and inherited private variants in MAP1B in periventricular nodular heterotopia. PLoS Genet. 2018;14(5):e1007281.
[8] Epilepsy Phenome/Genome Project, Epi4k Consortium. Diverse genetic causes of polymicrogyria with epilepsy. Epilepsia. 2021;62(4):973-983.




