视神经损伤
-
Figure 1|Dynamic Notch signaling during retinal regeneration.
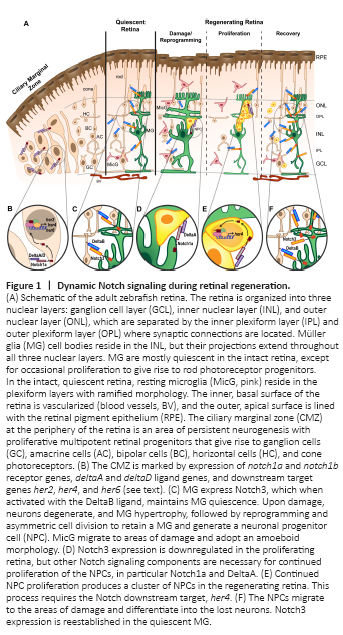
The ultimate strategy is to replace the missing neurons in the damaged retina and restore a healthy, functioning retina. Zebrafish (Danio rerio) possess the ability to regenerate lost neurons from an endogenous source, the Müller glia (Figure 1A). Unlike in mammals, adult zebrafish Müller glia respond to retinal damage by transitioning to a transient gliotic state before entering a reprogrammed state (Thomas et al., 2016; Hoang et al., 2020). A reprogrammed Müller glial cell divides asymmetrically to maintain the Müller glia and to produce a neuronal progenitor cell (NPC) (Nagashima et al., 2013; Lahne et al., 2015). The NPC continues to proliferate to produce a cluster of multipotent progenitors that differentiate into all retinal cell types with a bias towards the cells lost to damage (Powell et al., 2016; Lahne et al., 2021). The regenerated neurons develop normal morphologies, form stereotypical connections, and restore circuitry for visual function (D’Orazi et al., 2016; McGinn et al., 2018, 2019).
Notch receptor and ligand expression patterns influence the Notch regulation of cell fate by lateral specification (or lateral inhibition), which is the mechanism by which the fate of one cell influences the fate of its neighboring cells (Bray, 2016). This is accomplished through a negative feedback loop where the ligand-expressing cell induces the receptor-expressing cell to express Notch target genes, which are transcriptional repressors of the ligand genes, thereby repressing ligand expression in the signal-receiving cell. Within the damaged and regenerating retina, the mechanism of lateral specification may indicate to surviving cells, in particular the Müller glia, the state of the neighboring cells, as well as direct the fate of Müller glia-derived progenitor cells. Notch3 protein, which is expressed in Müller glia of the undamaged retina, is downregulated as Müller glia proliferate, and is reestablished in Müller glia of the recovering retina (Campbell et al., 2021). Alternatively, deltab mRNA is expressed throughout the undamaged retina and more highly in neurons than in the Müller glia, suggesting a complementary pattern to notch3 expression (Campbell et al., 2021). DeltaB-Notch3 interaction is required to maintain the Müller glia in a quiescent, non-proliferative state in the intact retina (Figure 1C and F; Campbell, et al., 2021). In addition, expression of dll4 is downregulated following injury and can be increased by forced expression of Fgf8a or Tgfb3, both of which inhibit injury-dependent proliferation, suggesting that Dll4 may also be a candidate ligand for Notch3 (Wan and Goldman, 2017; Lee et al., 2020). However, morpholino-mediated knockdown of Dll4 expression reduced Müller glia proliferation during light damage unlike the knockdowns of Notch3 and DeltaB, which both increased proliferation during light damage (Campbell et al., 2021).
Alternatively, the deltaA and deltaD ligands and the notch1a receptor are expressed in the developing zebrafish retina with expression gradually restricted over time to the ciliary marginal zone (Taylor et al., 2015), an area of persistent neurogenesis throughout the life of the fish (Figure 1B) (Hitchcock et al., 2004). Data from single-cell RNA-seq studies suggest that, similar to the developing retina, deltaA, deltaD, and notch1a are expressed in reprogrammed/activated Müller glia at the time of peak Müller glia proliferation (36 hours light treatment), with deltaA, deltaC, and deltaD also expressed in Müller glia-derived progenitors (Figure 1D; Hoang et al., 2020). Since Notch1 activity is known to maintain retinal progenitors in a proliferative state in the developing retina (Jadhav et al., 2006a), then the outcomes of Notch activation during regeneration may be dictated, in part, by specific ligand-receptor expression patterns.
Figure 2|Core components of the Notch signaling pathway.
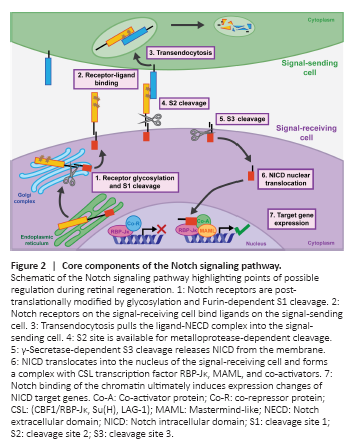
A complex network of signaling pathways is necessary as retinal damage breaks cell-cell contacts and new contacts are formed through remodeling, proliferation, and differentiation. Notch signaling is an obvious candidate as a major regulator of retinal regeneration because of its well-characterized role in retinal progenitor cell maintenance and Müller glia specification during development (Furukawa et al., 2000; Jadhav et al., 2006a, b). Additionally, Notch signaling relies on contact-mediated interactions between neighboring cells, which means the loss of cell contacts during retinal damage and the generation of different cell contacts as NPCs are generated, proliferate, and commit to neuronal fates are opportunities for Notch involvement. Notch signaling is activated when the Notch receptor expressed on the signal-receiving cell binds a ligand expressed on the signal-sending cell. A series of proteolytic cleavages are induced to release the Notch intracellular domain (NICD) for translocation into the nucleus of the signal-receiving cell where it effects gene expression changes (Henrique and Schweisguth, 2019). The outcomes of Notch signaling are highly context dependent with respect to specific tissue and to time, so despite the absence of signaling intermediates or second messenger amplification, there are abundant modes of Notch signaling regulation. These include post-translational modification during receptor production, ligand-receptor expression patterning, transendocytosis within the signal-sending cell, receptor cleavage events, NICD-dependent regulation of target gene transcription, and interactions with other signaling pathways (Figure 2).