NRR:安徽师范大学李君团队发现东方蝾螈非凡脊髓损伤修复能力的基因基础
撰写:王迪,赵曼,汤枭,高曼,刘文静,向明慧,阮健,陈杰,龙滨,李君
虽然人类等哺乳动物的脊髓再生能力有限,但同为脊索动物,海鞘[1]、斑马鱼[2]、非洲爪蟾[3]、壁虎[4]、蝾螈[5]、蜥蜴[6]等均有脊髓再生能力,其中四足动物蝾螈表现出最广泛的再生能力,其可再生四肢、尾巴、眼睛、心脏和中枢神经系统[7],是一种经典的再生模型。
来自中国安徽师范大学李君博士团队在《中国神经再生研究(英文版)》(Neural Regeneration Research)上发表了题为“Transcriptomic analysis of spinal cord regeneration after injury in Cynops orientalis”的研究,发现东方蝾螈(Cynops Orientalis)具有非凡的脊髓损伤修复能力,其脊髓横断后可自行恢复,且成熟神经元数量在损伤后能快速增加,其可在3周左右恢复了大部分运动功能,且整个过程未见胶质瘢痕产生。转录组测序筛选出13,059种差异表达的基因。这一结果为揭示脊髓损伤修复机制奠定了基础。
蝾螈的再生能力极强,其一生均存在修复受损脊髓的能力[8],如果能够探明蝾螈脊髓再生的分子机制,将为实现哺乳动物脊髓再生研究提供思路。李君等选取在中国广泛分布的东方蝾螈作为实验对象,从构建脊髓损伤模型开始,通过组织化学和行为学方法研究了脊髓损伤修复的过程,通过对假手术组以及损伤4和10d时的脊髓进行转录组测序筛选出脊髓损伤不同时期的差异基因表达谱,最后分析了可能参与脊髓损伤和修复的信号通路和相关基因。
东方蝾螈在脊髓损伤4和10d时可见明显的结缔组织再生,其受损的脊髓被逐渐修复(图1),排列在中央管周围遭到破坏的细胞也可自主修复(图2)。损伤后,神经元数量增加,损伤后第4天时胶质纤维酸性蛋白阳性的细胞数量增加,而白质中NeuN阳性的成熟神经元的数量在4和10d时均显著增加(图2)。
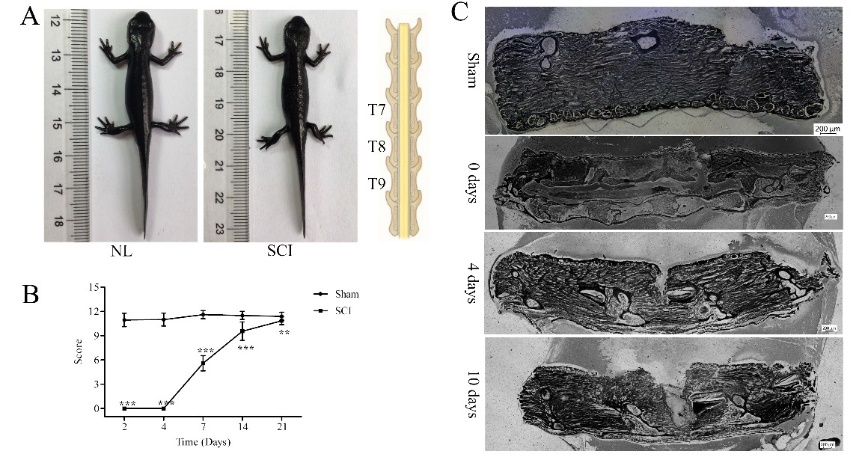
图1东方蝾螈脊髓损伤模型的建立(图源:Wang et al., Neural Regen Res, 2023)
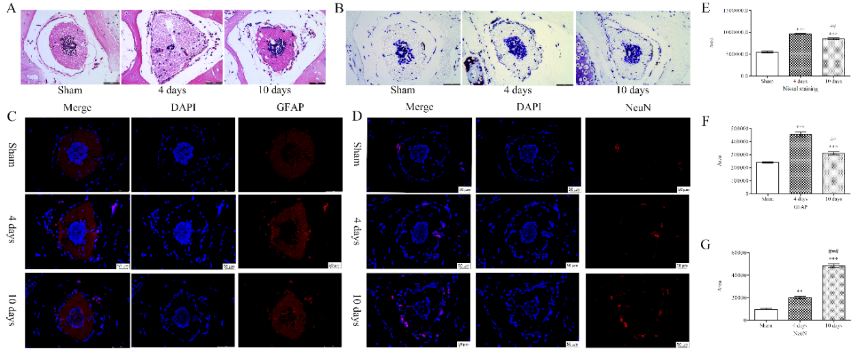
图2东方蝾螈脊髓损伤后细胞形态的变化(图源:Wang et al., Neural Regen Res, 2023)
转录组测序筛选出13,059种差异表达的基因(图3),这些基因可归入8个不同趋势表达谱(profile),其中4886种(37.31%)基因在表达谱6和7中;6537种(50.06%)基因在表达谱0和1中(图4);表达谱0,1,6和7中的基因相互之间有很大的不同。表达谱6有3322种基因,其在脊髓损伤后第4天明显升高,在10d时保持不变;与假手术组相比,4和10d时基因表达水平更高;表达谱7中的1564种基因在4和10d时均持续增加,其中4d时的表达水平明显高于假手术组,且10d时的表达水平高于4d,这意味着这些基因对蝾螈从脊髓损伤中恢复的能力至关重要。因此,蝾螈的脊髓损伤再生可能是由表达谱6和 7的基因介导的。
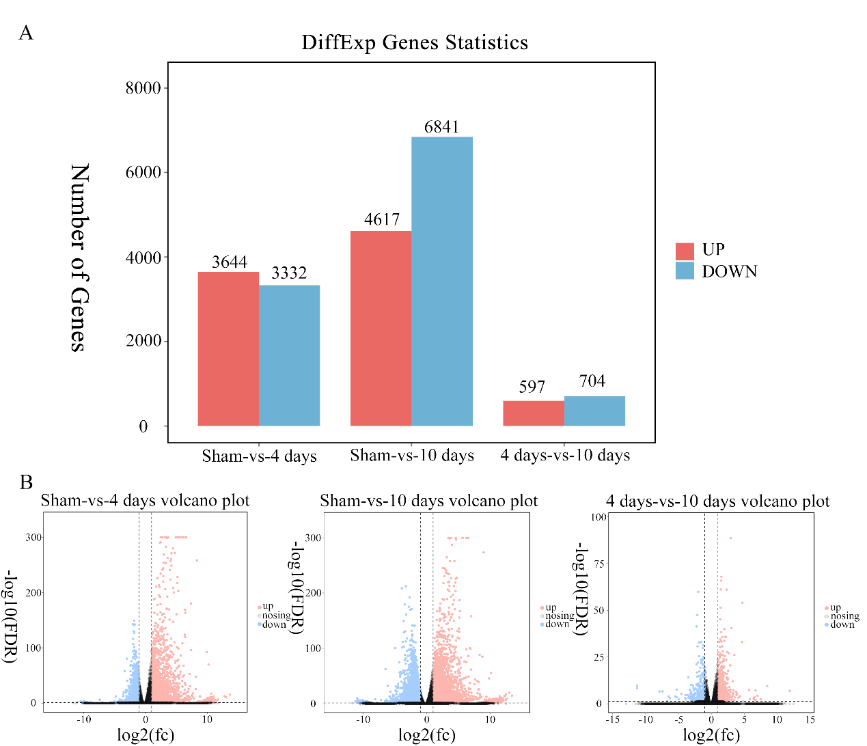
图3东方蝾螈脊髓损伤后不同时间的差异表达基因(图源:Wang et al., Neural Regen Res, 2023)

图4东方蝾螈脊髓损伤后差异表达基因的趋势分析(图源:Wang et al., Neural Regen Res, 2023)
在整个修复过程中,差异基因富集的特定KEGG信号通路包括细胞因子-细胞因子受体相互作用、神经活性配体1、神经活性配体-受体相互作用、病毒蛋白-细胞因子和细胞因子受体相互作用、肿瘤坏死因子信号通路、硫酸角蛋白的糖胺聚糖生物合成、硫酸角蛋白的糖胺聚糖生物合成等(图5)。
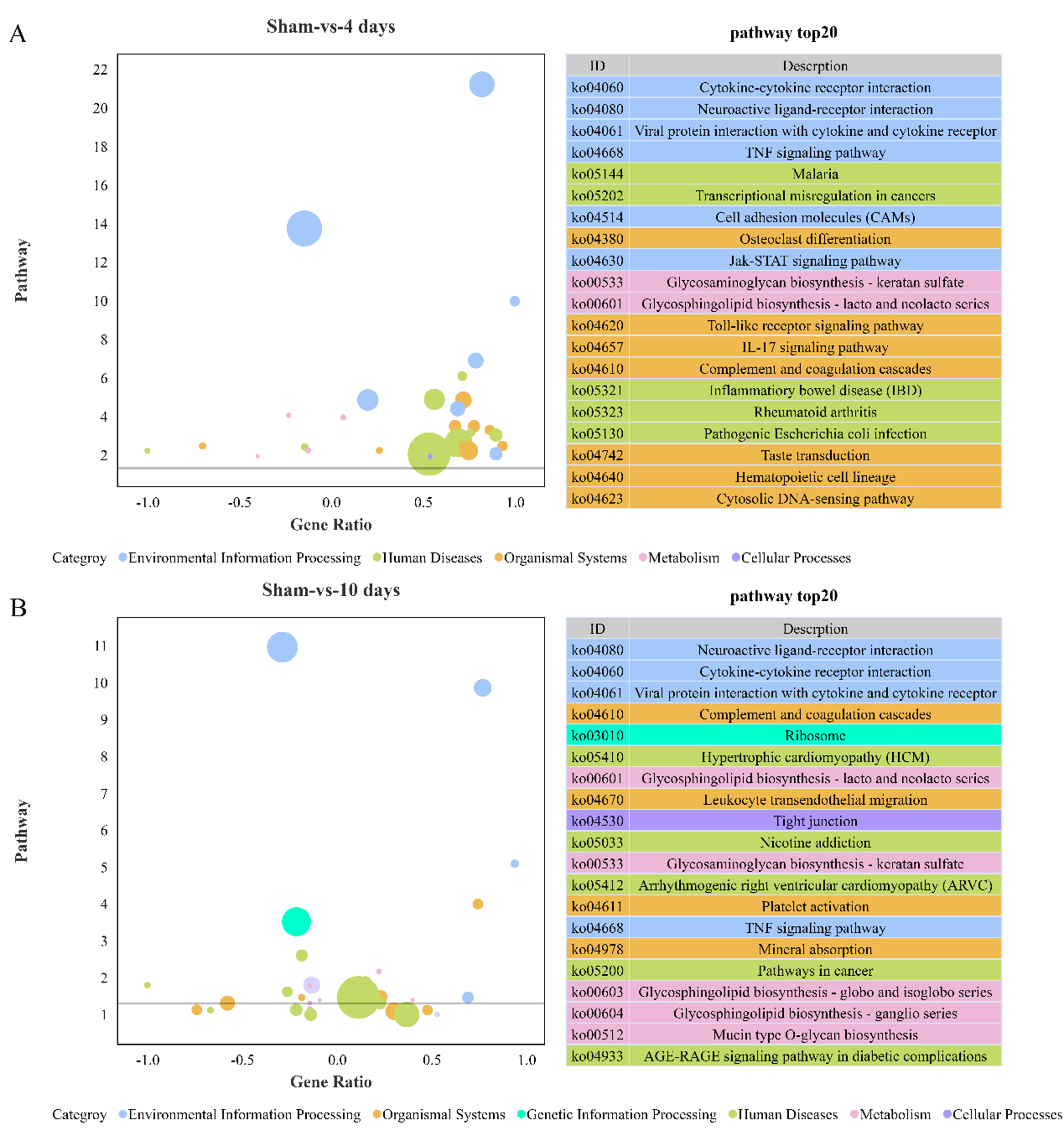
图5东方蝾螈脊髓损伤后差异表达基因的KEGG富集分析(图源:Wang et al., Neural Regen Res, 2023)
将差异表达基因与哺乳动物脊髓损伤后响应的信号通路中的基因对比,发现上调的基因包括FOS、Egr1、Zfp36、Btg2、FKBP3、Fkbp1a、GAP43、Gfap、VIM、XYLT1、Col4a1、VCAN;下调的基因则包括Nr4a1、SOX9、SEMA6A、Plxna2、CHST11、Col2a1、Cspg4、NCAN、Ptprz1、ACAN、BCAN(图6),这些差异表达基因为进一步研究其在脊髓损伤及修复中作用提供理论依据。
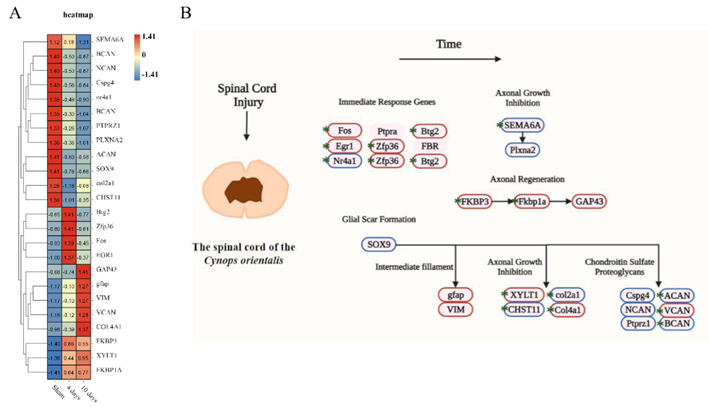
图 6 脊髓损伤相关信号通路中基因表达的变化(图源:Wang et al., Neural Regen Res, 2023)
该研究认为,作为中国南方常见的有尾两栖类,东方蝾螈的差异表达基因可为研究其在脊髓损伤及修复中作用提供帮助。
原文链接:https://doi.org/10.4103/1673-5374.373717
参考文献
[1] Jiang D, Munro EM, Smith WC. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr Biol. 2005;15(1):79-85.
[2] Briona LK, Dorsky RI. Spinal cord transection in the larval zebrafish. J Vis Exp. 2014(87):51479.
[3] Forehand CJ, Farel PB. Anatomical and behavioral recovery from the effects of spinal cord transection: dependence on metamorphosis in anuran larvae. J Neurosci. 1982;2(5):654-652.
[4] Xu M, Wang T, Li W, et al. PGE2 facilitates tail regeneration via activation of Wnt signaling in Gekko japonicus. J Mol Histol. 2019;50(6):551-562.
[5] Fior J. Salamander regeneration as a model for developing novel regenerative and anticancer therapies. J Cancer. 2014;5(8):715-719.
[6] Simpson SB, Jr. Analysis of tail regeneration in the lizard lygosoma laterale. I. Initiation of regeneration and cartilage differentiation: the role of ependyma. J Morphol. 1964;114:425-435.
[7] Carducci F, Carotti E, Gerdol M, et al. Investigation of the activity of transposable elements and genes involved in their silencing in the newt Cynops orientalis, a species with a giant genome. Sci Rep. 2021;11(1):14743.
[8] Donovan J, Kirshblum S. Clinical trials in traumatic spinal cord injury. Neurotherapeutics. 2018;15(3):654-668.
第一作者:王迪,1996年生,女,安徽师范大学2019级硕士。
通讯作者:李君,安徽师范大学生理学与发育生物学课题组。





